Proteomics Data Analysis and Visualization
Unlocking Biological Insights: A Comprehensive Guide to Proteomics Data Analysis and Visualization
3/6/20254 min read
Introduction to Proteomics
Proteomics is a critical branch of molecular biology that focuses on the large-scale study of proteins, particularly their functions and structures. Given that proteins are pivotal to numerous biological processes, understanding them provides significant insights into the molecular underpinnings of life. By analyzing the complete set of proteins, or proteome, expressed by a cell, tissue, or organism at a specific time, researchers can unveil intricate biological mechanisms and pathways that underlie health and disease.
The field of proteomics is multifaceted, encompassing several types of studies, including comparative, quantitative, and functional proteomics. Comparative proteomics involves assessing differences in protein expression between various biological states, such as healthy and diseased tissue. This approach is invaluable for identifying biomarkers and potential therapeutic targets. Quantitative proteomics aims to determine the concentration of proteins under different conditions, thus providing insights into cellular responses and regulatory mechanisms. On the other hand, functional proteomics focuses on understanding protein functions and interactions, elucidating how proteins collaborate in complex biological systems.
Recent advancements in technologies such as mass spectrometry and bioinformatics have dramatically transformed proteomics. Mass spectrometry allows for the precise identification and quantification of proteins, enabling researchers to analyze complex biological samples with unprecedented accuracy. Concurrently, bioinformatics tools facilitate the management and interpretation of vast datasets generated through proteomics studies, marking a significant leap in the field's analytical capabilities. The integration of these technologies not only enhances the resolution and depth of proteomic analyses but also accelerates discoveries that could lead to innovative therapeutic interventions.
As we delve further into the realms of data analysis and visualization, the foundational understanding of proteomics will be essential for harnessing the full potential of protein-related insights in biological research.
Key Techniques in Proteomics Data Analysis
Proteomics data analysis involves a range of methodologies designed to explore the protein composition within biological samples. Among these techniques, mass spectrometry (MS) stands out as a pivotal tool. Mass spectrometry offers high sensitivity and specificity, enabling the identification and quantification of proteins in complex mixtures. This technique works by ionizing protein fragments and then measuring their mass-to-charge ratios, thereby producing a spectra that can be interpreted to deduce protein identity and abundance. Additionally, MS can provide valuable insights into post-translational modifications, crucial for understanding protein functionality.
In tandem with mass spectrometry, gel-based techniques, specifically two-dimensional gel electrophoresis (2-DE), remain significant in the field. This method separates proteins based on their isoelectric points and molecular weights, allowing for a detailed analysis of protein expression patterns within different biological conditions. The resolved protein spots can be further analyzed using mass spectrometry for identification, making this a powerful approach in proteomics.
Another innovative technique includes the use of protein microarrays, facilitating the simultaneous measurement of thousands of proteins. This method involves immobilizing proteins onto a solid surface, where binding interactions can be measured in real-time. Protein microarrays are particularly useful for biomarker discovery and for understanding protein-protein interactions in a high-throughput manner.
The data processing steps are crucial in proteomics analysis. Protein identification and quantification typically require robust computational tools and algorithms to ensure accurate results. The application of advanced statistical methods helps in interpreting the data, identifying significant changes in protein levels across different samples. Furthermore, employing suitable software tools and databases is essential for effective data management, enabling researchers to integrate and analyze datasets efficiently. Thus, a well-structured analysis workflow incorporating these techniques is fundamental for unlocking biological insights through proteomics.
Visualizing Proteomics Data: Tools and Techniques
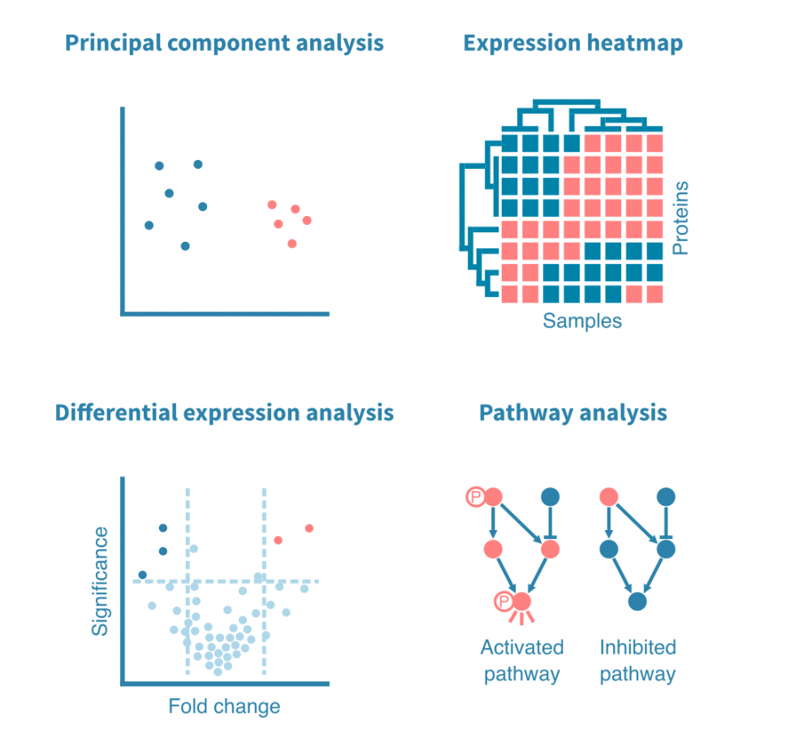
Effective data visualization serves as a cornerstone in the field of proteomics research, enabling researchers to interpret complex datasets and discern meaningful biological insights. Visualizations offer an accessible means to represent information, facilitating communication among scientists and aiding in decision-making processes. Commonly used visualizations in proteomics include heat maps, volcano plots, and protein interaction networks, each with distinct applications and strengths.
Heat maps are particularly effective in displaying the expression levels of proteins across different samples. By using color gradients, heat maps allow researchers to quickly identify patterns, such as upregulated or downregulated proteins under specific conditions. These visualizations are instrumental in identifying trends and anomalies within large datasets, ultimately aiding in the understanding of biological responses to various stimuli.
Volcano plots represent another critical visualization technique, integrating fold-change and statistical significance in a single graphical format. This method allows researchers to delineate significant changes in protein expression, making it easier to highlight potential biomarkers or therapeutic targets. By plotting p-values against fold-change, the plot's pronounced features can direct attention to proteins of interest, streamlining the analysis process.
Protein interaction networks provide a holistic view of the interconnectedness within protein complexes and pathways. These visualizations depict how proteins interact with one another, revealing potential regulatory mechanisms and signaling cascades. Tools such as Cytoscape allow researchers to visualize and analyze these networks, offering insights into the functional roles of proteins within biological systems.
Several software packages are available for creating these visualizations, including R and Python-based libraries, which provide flexibility and customization options. While these tools offer impressive capabilities, each comes with its own advantages and limitations concerning ease of use, specific features, and the learning curve associated with them. Ultimately, the choice of visualization tool depends on the research context and the desired presentation format.
Future Trends and Challenges in Proteomics
The field of proteomics is rapidly evolving, with significant advancements on the horizon. One of the most promising trends involves single-cell proteomics, which aims to unravel the complexities of protein expression at the individual cell level. This innovative approach enables researchers to gain insights into cellular heterogeneity, which is crucial for understanding disease mechanisms and tailoring personalized therapies. As technology progresses, single-cell proteomics is becoming more accessible, facilitating detailed studies that could lead to breakthroughs in cancer research, neurodegenerative diseases, and other health conditions.
Moreover, the integration of proteomics with other 'omics' disciplines, such as genomics and metabolomics, is gaining traction. This multi-omics approach provides a comprehensive understanding of biological systems by combining data from various layers of cellular regulation. Researchers are now focusing on creating integrative frameworks and innovative analytical tools to effectively combine and interpret complex data sets. Such synergies have the potential to advance precision medicine by illuminating the interconnected pathways that drive disease processes.
However, while these advancements are promising, there are several challenges that need to be addressed. The complexity of proteomics data presents a significant hurdle, as the sheer volume and variability of data can complicate analyses and interpretation. Reproducibility remains a critical concern, with variations in sample preparation, instrument calibration, and analytical techniques often leading to inconsistent results across studies. Additionally, the lack of standardized protocols and common data formats further complicates collaborative research efforts.
In conclusion, it is essential for researchers to embrace innovative techniques and prioritize collaborative initiatives to overcome these challenges. By doing so, the field of proteomics can advance more effectively, ultimately leading to impactful discoveries that enhance our understanding of biological systems and improve human health.
